Loading FCMs into R for fcmconfr
Types of FCMs
When people create fuzzy cognitive maps (FCMs) they are asked to provide weights for all edges between causally connected concepts in the map.
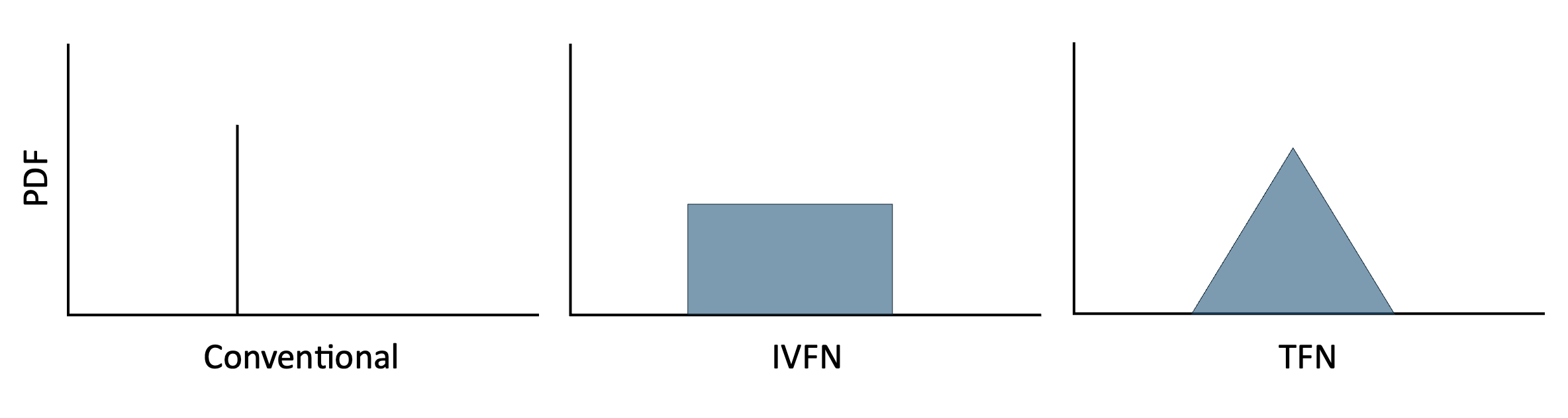
In conventional FCMs, edge weights are individual values (e.g., -0.5, 0.25, etc.) that express a relationship between two concepts without uncertainty. However, FCMs can also describe edge weights using (1) Interval-Value Fuzzy Numbers (IVFNs) where weights are expressed as ranges and no one value is more likely than another (e.g., [min, max]) or (2) Triangular Fuzzy Numbers (TFNs) where weights are expressed as ranges, but with values more likely to be near a particular value within those ranges (e.g., [min, mode, max]).
The figure below illustrates the probability density functions
associated with. each type of edge weight. fcmconfr can
accomodate FCMs with all three edge weight types. This vignette
illustrates the workflow for preparing and importing these FCMs into R
and analysis with fcmconfr.
1. Conventional FCMs
We recommend storing conventional FCM adjacency matrices in excel workbooks or .csv files and importing them into R.
Weights are read from row to column (i.e., a value of 0.25 in row 2, column 3 indicates that increasing the node in row 2 is perceived to increase the node in column 3 by 0.25). The examples below show how to load adjacency matrices from .xlsx files. A similar process is used for .csv, .xml or other file types.
Note: Use file.choose() to get the path to a file
you want to import. There is not a similar function to get the path to a
directory, but users may extrapolate from the output of
file.choose() as needed.
# For an individual FCM adjacency matrix
adj_matrix <- readxl::read_excel(filepath)1.1 - Conventional FCMs from Different Sheets in a Single .xlsx File
# For multiple FCM adjacency matrices stored in different sheets in the same .xlsx file
sheets <- readxl::excel_sheets(filepath)
adj_matrices <- lapply(
sheets,
function(sheet) readxl::read_excel(filepath, sheet = sheet)
)
# For multiple FCM adjacency matrices stored in separate .xlsx files in the
# same directory
filepaths <- list.files(directory_path, full.names = TRUE)
adj_matrices <- lapply(
filepaths,
function(filepath) readxl::read_excel(filepath)
)2. IVFN FCMs
IVFN FCMs are more complicated data objects because each edge weight is described by two values (an upper and a lower bound) and common file formats like .xlsx and .csv can only store one value per cell.
We recommend importing two separate adjacency matrices for each
IVFN-FCM, one that contains only lower bound edge weights and one that
contains only upper bound edge weights. Once these matrices have been
imported, the fcmconfr function
make_adj_matrix_w_ivfns() can be used to create a single
IVFN-FCM adjacency matrix.
2.1 - Loading an IVFN-FCM from a single .xlsx file:
To load an IVFN-FCM from a single .xlsx file, the file must contain two sheets, one for the lower bound adjacency matrix and one for the upper bound adjacency matrix. See example below:
sheets <- readxl::excel_sheets(filepath)
# If lower and upper adjacency matrices are in the same .xlsx file across two different sheets
lower_and_upper_adj_matrices <- lapply(
sheets,
function(sheet) readxl::read_excel(filepath, sheet = sheet)
)
lower_adj_matrix <- lower_and_upper_adj_matrices[[1]]
upper_adj_matrix <- lower_and_upper_adj_matrices[[2]]
ivfn_adj_matrix <- make_adj_matrix_w_ivfns(lower_adj_matrix, upper_adj_matrix)2.2 - Loading an IVFN-FCM from Separate Files in the Same Directory
This method must be used if loading data from separate .xlsx, .csv, etc., files.
# If lower and upper adjacency matrices are in separate .xlsx files
lower_adj_matrix_filepath <- file.path(directory_path, "fcm_1_lower.xlsx") # Your filename here
upper_adj_matrix_filepath <- file.path(directory_path, "fcm_1_upper.xlsx") # Your filename here
lower_adj_matrix <- readxl::read_excel(lower_adj_matrix_filepath)
upper_adj_matrix <- readxl::read_excel(upper_adj_matrix_filepath)
ivfn_adj_matrix <- make_adj_matrix_w_ivfns(lower_adj_matrix, upper_adj_matrix)2.3 - Loading Multiple IVFN-FCMs from Separate Files in the Same Directory
Use this method when loading a set IVFN-FCMs in the same directory from .xlsx files with both the lower and upper bound adjacency matrices stored in the same file.
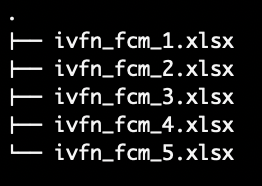
ivfn_fcm_1.xlsx has a sheet for the lower bound adjacency
matrix and another sheet for the upper bound adjacency matrix.Load the following function into the Global Environment to run the
example workflow in fewer lines of code or repetitively in a
for loop or using lapply().
# Load into Global Environment to streamline IVFN FCM imports from individual .xlsx files
import_ivfn_fcm_from_single_xlsx <- function(filepath) {
sheets <- readxl::excel_sheets(filepath)
lower_and_upper_adj_matrices <- lapply(
sheets,
function(sheet) readxl::read_excel(filepath, sheet = sheet)
)
ivfn_adj_matrix <- make_adj_matrix_w_ivfns(
lower_and_upper_adj_matrices[[1]], lower_and_upper_adj_matrices[[2]]
)
ivfn_adj_matrix
}Then, call the newly-defined
import_ivfn_fcm_from_single_xlsx function within
lapply or a loop.
# For multiple IVFN FCM adjacency matrices stored in separate .xlsx files stored in the
# same directory
filepaths <- list.files(directory_path, full.names = TRUE)
ivfn_adj_matrices <- lapply(
filepaths,
function(filepath) import_ivfn_fcm_from_single_xlsx(filepath)
)2.4 - Loading Multiple IVFN-FCMs from Separate Files in Different Directories
Use this method when loading a set of IVFN-FCMs where the lower and upper bound adjacency matrices for a single IVFN-FCM are stored in separate files.
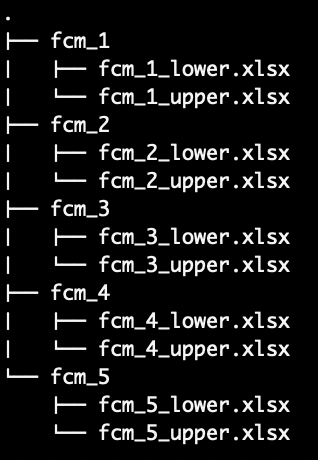
fcm_1, fcm_2, etc.,) with each sub-directory
containing separate .xlsx files for the lower and upper adjacency
matrices. For example, the an IVFN-FCM can be created from the
fcm_1 sub-directory with fcm_1_lower.xlsx and
fcm_1_upper.xlsx containing data for the lower and upper
bound adjacency matrices respectively.Load the following function into the Global Environment to run the
example workflow in fewer lines of code or repetitively in a
for loop or using lapply().
# Load into Global Environment to streamline IVFN FCM imports from two .xlsx files
import_ivfn_fcm_from_two_xlsx_files <- function(lower_adj_matrix_filepath,
upper_adj_matrix_filepath) {
# Use read.csv or similar for other file types
lower_adj_matrix <- readxl::read_excel(lower_adj_matrix_filepath)
upper_adj_matrix <- readxl::read_excel(upper_adj_matrix_filepath)
ivfn_adj_matrix <- make_adj_matrix_w_ivfns(lower_adj_matrix, upper_adj_matrix)
ivfn_adj_matrix
}Then, call the newly-defined
import_ivfn_fcm_from_two_xlsx_files function within
lapply or a loop.
# For IVFN FCM adjacency matrices stored in separate .xlsx files across different directories
directory_paths <- file.path(directory_path, list.files(directory_path)) # Note list.files shows dirs here
ivfn_adj_matrices <- lapply(
directory_paths,
function(directory_path) {
ivfn_files <- list.files(directory_path)
lower_ivfn_adj_matrix_file <- ivfn_files[[1]] # Order as needed or by filename
upper_ivfn_adj_matrix_file <- ivfn_files[[2]] # Order as needed or by filename
lower_ivfn_adj_matrix_filepath <- file.path(directory_path, lower_ivfn_adj_matrix_file)
upper_ivfn_adj_matrix_filepath <- file.path(directory_path, upper_ivfn_adj_matrix_file)
imported_ivfn_adj_matrices <- import_ivfn_fcm_from_two_xlsx_files(
lower_ivfn_adj_matrix_filepath,
upper_ivfn_adj_matrix_filepath
)
imported_ivfn_adj_matrices
}
)3. TFN FCMs
TFN FCMs are the most complicated edge weights because they are
described by three values, an upper bound, a lower bound and a mode that
indicates the most-likely value for the edge. TFN-FCMs are imported
using the same general approach as IVFN-FCMs, but require 3 adjacency
matrices rather than 2. Once the three adjacency matrices have been
imported, the fcmconfr function
make_adj_matrix_w_tfns() can be used to create a single
TFN-FCM adjacency matrix.
3.1 - Loading a TFN-FCM from a Single .xlsx File
To load a TFN-FCM from a single .xlsx file, the file must contain three sheets, one for the lower bound adjacency matrix, one for the upper bound adjacency matrix, and another for the mode adjacency matrix.
sheets <- readxl::excel_sheets(filepath)
# If lower, upper and mode adjacency matrices are in the
# same .xlsx file across 3 different sheets
lower_mode_and_upper_adj_matrices <- lapply(
sheets,
function(sheet) readxl::read_excel(filepath, sheet = sheet)
)
lower_adj_matrix <- lower_mode_and_upper_adj_matrices[[1]]
mode_adj_matrix <- lower_mode_and_upper_adj_matrices[[2]]
upper_adj_matrix <- lower_mode_and_upper_adj_matrices[[3]]
tfn_adj_matrix <- make_adj_matrix_w_tfns(lower_adj_matrix, mode_adj_matrix, upper_adj_matrix)3.2 - Loading a TFN-FCM from Separate Files in the Same Directory
Use this method if loading data from separate .xlsx, .csv, etc., files.
# If lower, mode, and upper adjacency matrices are in separate .xlsx files
lower_adj_matrix_filepath <- file.path(directory_path, "fcm_1_lower.xlsx") # Your filename here
mode_adj_matrix_filepath <- file.path(directory_path, "fcm_1_mode.xlsx") # Your filename here
upper_adj_matrix_filepath <- file.path(directory_path, "fcm_1_upper.xlsx") # Your filename here
lower_adj_matrix <- readxl::read_excel(lower_adj_matrix_filepath)
mode_adj_matrix <- readxl::read_excel(mode_adj_matrix_filepath)
upper_adj_matrix <- readxl::read_excel(upper_adj_matrix_filepath)
tfn_adj_matrix <- make_adj_matrix_w_tfns(lower_adj_matrix, mode_adj_matrix, upper_adj_matrix)3.2 - TFN FCMs from a Directory of Adj. Matrices in Single .xlsx Files
Use this method when loading a set TFN-FCMs in the same directory from .xlsx files with the lower, mode and upper bound adjacency matrices stored in the same file.
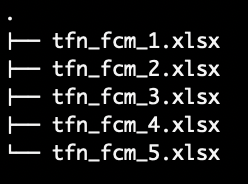
tfn_fcm_1.xlsx has a sheet for the lower bound adjacency
matrix, the mode adjacency matrix, and another sheet for the upper bound
adjacency matrix.Load the following function into the Global Environment to run the
example workflow in fewer lines of code or repetitively in a
for loop or using lapply().
# Load into Global Environment to streamline TFN FCM imports from individual .xlsx files
import_tfn_fcm_from_single_xlsx <- function(filepath) {
sheets <- readxl::excel_sheets(filepath)
lower_mode_and_upper_adj_matrices <- lapply(
sheets,
function(sheet) readxl::read_excel(filepath, sheet = sheet)
)
tfn_adj_matrix <- make_adj_matrix_w_tfns(
lower_mode_and_upper_adj_matrices[[1]],
lower_mode_and_upper_adj_matrices[[2]],
lower_mode_and_upper_adj_matrices[[3]]
)
tfn_adj_matrix
}Then, call the newly-defined
import_tfn_fcm_from_single_xlsx function within
lapply or a loop.
# For multiple TFN FCM adjacency matrices stored in
# separate .xlsx files stored in the same directory
filepaths <- list.files(directory_path, full.names = TRUE)
tfn_adj_matrices <- lapply(
filepaths,
function(filepath) import_tfn_fcm_from_single_xlsx(filepath)
)3.3 - TFN FCMs from a Adj. Matrices in Separate .xlsx Files Stored in Different Directories
Use this method when loading a set of TFN-FCMs where the lower, mode and upper bound adjacency matrices for a single TFN-FCM are stored in separate files.
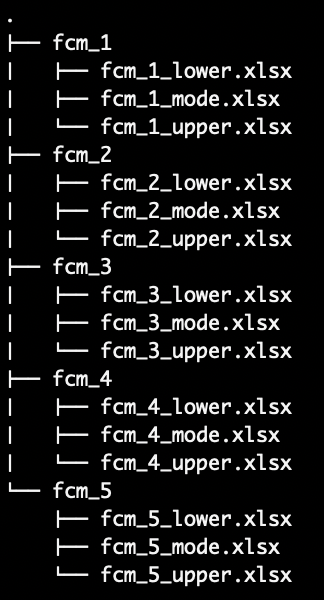
fcm_1, fcm_2, etc.,) with each sub-directory
containing separate .xlsx files for the lower, mode and upper adjacency
matrices. For example, a TFN-FCM can be created from the
fcm_1 sub-directory with fcm_1_lower.xlsx,
fcm_1_mode.xlsx and fcm_1_upper.xlsx
containing data for the lower, mode and upper bound adjacency matrices
respectively.Load the following function into the Global Environment to run the
example workflow in fewer lines of code or repetitively in a
for loop or using lapply().
# Load into Global Environment to streamline tFN FCM imports from two .xlsx files
import_tfn_fcm_from_two_xlsx_files <- function(lower_adj_matrix_filepath,
mode_adj_matrix_filepath,
upper_adj_matrix_filepath) {
# Use read.csv or similar for other file types
lower_adj_matrix <- readxl::read_excel(lower_adj_matrix_filepath)
mode_adj_matrix <- readxl::read_excel(mode_adj_matrix_filepath)
upper_adj_matrix <- readxl::read_excel(upper_adj_matrix_filepath)
tfn_adj_matrix <- make_adj_matrix_w_tfns(
lower_adj_matrix, mode_adj_matrix, upper_adj_matrix
)
tfn_adj_matrix
}Then, call the newly-defined
import_tfn_fcm_from_two_xlsx_files function within
lapply or a loop.
# For TFN FCM adjacency matrices stored in separate .xlsx files across different directories
directory_paths <- file.path(directory_path, list.files(directory_path)) # Note list.files shows dirs here
tfn_adj_matrices <- lapply(
directory_paths,
function(directory_path) {
tfn_files <- list.files(directory_path)
lower_tfn_adj_matrix_file <- tfn_files[[1]] # Order as needed or by filename
mode_tfn_adj_matrix_file <- tfn_files[[2]] # Order as needed or by filename
upper_tfn_adj_matrix_file <- tfn_files[[3]] # Order as needed or by filename
lower_tfn_adj_matrix_filepath <- file.path(directory_path, lower_tfn_adj_matrix_file)
mode_tfn_adj_matrix_filepath <- file.path(directory_path, mode_tfn_adj_matrix_file)
upper_tfn_adj_matrix_filepath <- file.path(directory_path, upper_tfn_adj_matrix_file)
imported_tfn_adj_matrices <- import_tfn_fcm_from_two_xlsx_files(
lower_tfn_adj_matrix_filepath,
mode_tfn_adj_matrix_filepath,
upper_tfn_adj_matrix_filepath
)
imported_tfn_adj_matrices
}
)